Merging Neurons from the FlyWire into main CATMAID#
Unlike for the Google segmentation, flywire does not have precomputed skeletons - which makes sense given that it is a changing data set. This means that we have to generate the skeletons ourselves.
Important
It is your responsibility to make sure that you have permission to import
given data from flywire into CATMAID. If you are unsure, please get in touch
with flywire’s community manager Claire McKellar. See also
fafbseg.flywire.fetch_edit_history() to figure out who else has been
working on a neuron of interest.
I highly recommend you familiarise yourself with the functions used in this example - we are using many default values here but depending on the neuron you are trying to import, you might need to adjust things.
Setting things up#
First things first: if you haven’t already please generate and save your chunkedgraph secret so that you can fetch flywire data.
Now fire up a Python terminal:
ipython
Import the relevant libraries:
import fafbseg
import pymaid
Next initialize the connection to main v14 CATMAID - make sure to replace
HTTP_USER, HTTP_PW and API_TOKEN with the corresponding credentials:
manual = pymaid.CatmaidInstance('https://neuropil.janelia.org/tracing/fafb/v14',
api_token='API_TOKEN',
http_user='HTTP_USER',
http_password='HTTP_PW',
caching=False,
max_threads=20)
Do the import#
OK now that we’re all set, we there is a single function that does the whole import for us:
# 720575940617303000 is the root ID of the neuron we want to import
resp = fafbseg.flywire.merge_flywire_neuron(id=720575940617303000,
target_instance=manual,
tag='YOURTAG',
flywire_dataset='production',
assert_id_match=True,
drop_soma_hairball=False)
The YOURTAG should be something that identifies you and/or your group. It
will be added as annotation to the neuron after merging.
This function will first skeletonize the flywire mesh and present the skeleton for your inspection. If accepted, it will proceed to merge the neuron into CATMAID. Carefully read the instructions printed in the terminal at each step!
Merge finished - What now?#
Success! The neuron has now been merged into existing manual tracings - what now?
Minimally you should have a look at the sites where existing and new
tracings were joined. The respective nodes will both be tagged
with Joined from/into {SKELETON_ID} and have a confidence of 1 so that they are
easy to find in the treenode table:
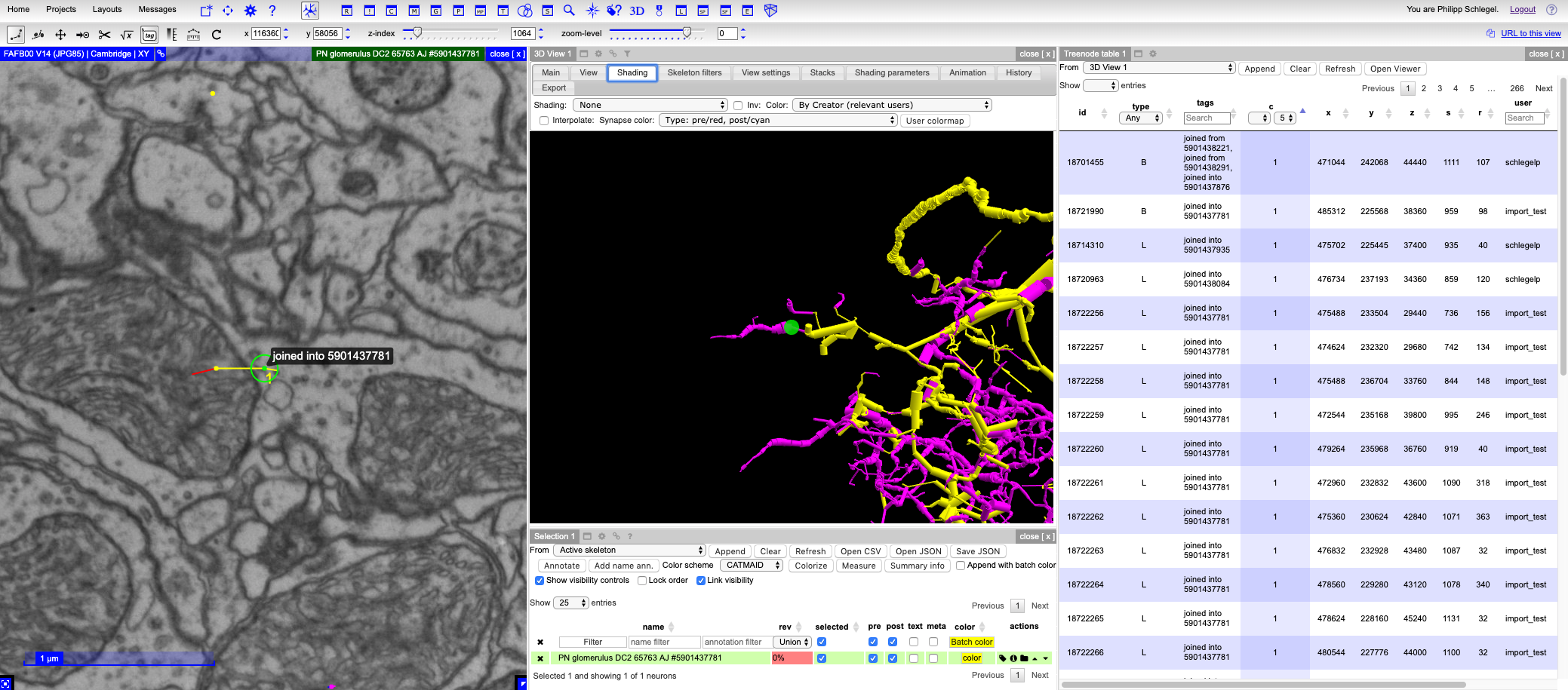
Depending on how much you care about the neuron, you want do a full review to make sure that nothing was missed during the merge process.
Caveats#
The merge procedure is a lengthy process and there is a chance that your local data will diverge from the live CATMAID server (i.e. people make changes that the script is unaware off). You should consider to:
upload neurons in only small batches (i.e. one at a time)
if possible make sure nobody is working on the neuron(s) you are merging into
ideally run the merge when few people in CATMAID are tracing
Something went wrong - What now?#
There are a few problems you might run into and that could cause the merging procedure to stop. Generally speaking, the script is failsafe: e.g. if the upload fails half-way through, you should be able to just restart and the script will recognise changes that have already been made and skip these.
Especially if you are on slow connections, you should consider decreasing the number of parallel requests allowed to lower the chances that something goes wrong:
# Default is 100 -> let's lower that
manual.max_threads = 20