Mirroring FlyWire neurons#
The FlyWire connectome revealed that the FAFB image dataset underlying it was accidentally flipped along the left-right (i.e. “x”) axis. A detailed explanation of this observation can be found in Schlegel et al.
Importantly, the official side labels we provide for FlyWire are biologically
correct. Let’s illustrate this:
import navis
import flybrains
import navis.interfaces.neuprint as neu
from fafbseg import flywire
import fafbseg.flywire.NeuronCriteria as NC
client = neu.Client("https://neuprint.janelia.org", dataset="hemibrain:v1.2.1")
# Fetch a couple neurons on the left side of the brain
# (note that "side" always refers to where the soma is)
meshes = flywire.get_mesh_neuron(
NC(type="DA1_lPN", side="left"), dataset="flat_630", lod=3
)
# Slightly downsample neurons for visualization
meshes = navis.downsample_neuron(meshes, 5)
Found 8 neurons matching the given criteria.
# Visualize the neurons
fig, ax = navis.plot2d(meshes, color="red")
# Add the neuropil
_ = navis.plot2d(flybrains.FLYWIRE, color=(0.8, 0.8, 0.8, 0.05), ax=ax)
ax.elev = -90 # rotate brain to frontal view
ax.set_box_aspect(None, zoom=0.7) # zoom out slightly
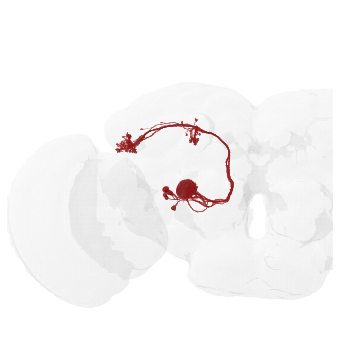
Note how the neurons appears on the left of the brain? Since this is a frontal view, left neurons should - by convention - appear on the right side of the image. But because of the aforementioned inversion of the FAFB image data what we’re seeing on the left here is the correct biological left of this fly’s brain! Or in other words: “image left” = “fly’s left”!
In many cases this may not actually matter but what if you are working on neurons or circuit that aren’t left/right symmetric - i.e. where laterality matters?
One of best studied examples of left-right asymmetry stems from two small neuropils close to the midline of the brain called the “asymmetric bodies” (AB). Here, the AB should be bigger on the fly’s right than on the left.
We can check what this should look like using the (non-flipped) hemibrain and by using the SA1-3 cell types which represent the main input to those neuropils:
sa13_hb = neu.fetch_mesh_neuron(neu.NeuronCriteria(type=("SA1", "SA2", "SA3")), lod=3)
fig, ax = navis.plot2d(sa13_hb, color="red")
_ = navis.plot2d(flybrains.JRCFIB2018Fraw, color=(0.8, 0.8, 0.8, 0.1), ax=ax)
ax.text3D(25909, 21760, 24709, "AB", c="red", ha="center")
ax.elev = -180 # rotate brain to frontal view
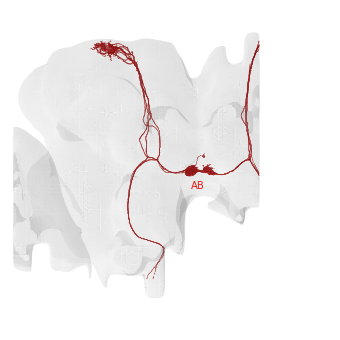
For the hemibrain (which is not inverted), “image left” = “fly’s right” and vice versa. As you can see the AB is substantially larger on the fly’s right than on the left.
Let’s try the same thing in FlyWire:
# Fetch SA1-3 cells in FlyWire
sa13 = flywire.search_annotations(
"hemibrain_type:SA[1-3]", materialization=630, regex=True
)
sa13_fw = flywire.get_mesh_neuron(sa13.root_id.values, dataset="flat_630", lod=3)
sa13_fw = navis.downsample_neuron(sa13_fw, 5) # slightly downsample for visualization
fig, ax = navis.plot2d(sa13_fw, color="red")
_ = navis.plot2d(flybrains.FLYWIRE, color=(0.8, 0.8, 0.8, 0.1), ax=ax)
ax.elev = -90 # rotate brain to frontal view
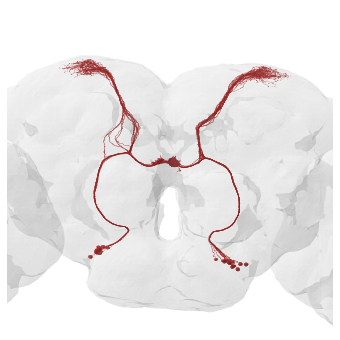
As expected, for FlyWire the AB on the right side of the image is larger. Keep in mind that “image right” = “fly’s right”!
What if we want if we want to compare the flipped FAFB/FlyWire neurons with non-inverted data such as the hemibrain? Easy: we just apply a non-rigid mirror transform to flip the data to its correct orientation:
# Mirror neurons left/right
sa13_fw_mirr = navis.mirror_brain(sa13_fw, template="FLYWIRE")
# Plot mirrored neurons in red
fig, ax = navis.plot2d(sa13_fw_mirr, color="r")
_ = navis.plot2d(flybrains.FLYWIRE, color=(0.8, 0.8, 0.8, 0.1), ax=ax)
ax.elev = -90 # rotate brain to frontal view
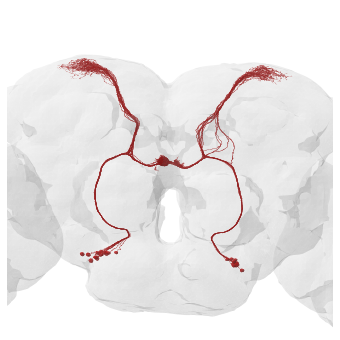
With the FlyWire neurons mirrored they now match the hemibrain, i.e. “image left” = “fly’s right”.