Fetching neurons#
In this tutorial you will learn to fetch various representations (meshes, skeletons and dotprops) for FlyWire neurons.
First up: imports!
>>> import navis
>>> from fafbseg import flywire
We will run our tutorial using the public dataset. To make our life easier we will set the default dataset:
>>> flywire.set_default_dataset("public")
Default dataset set to "public"
OK, with that out of the way let’s load some neurons:
>>> n = flywire.get_mesh_neuron(720575940625431866)
>>> n
| type | navis.MeshNeuron |
|---|---|
| name | None |
| id | 720575940625431866 |
| units | 1 nanometer |
| n_vertices | 309801 |
| n_faces | 622597 |
Now we can use navis to plot the neuron:
>>> fig, ax = navis.plot2d(n, color="red")
>>> ax.azim = ax.elev = -90

Note that FlyWire neurons are very detailed: the above neuron has over half a
million faces! That makes working with them slow and it’s a good idea to
downsample them if possible. You can use navis.downsample_neuron to reduce
the number of faces in a neuron
Alternatively, you can use skeleton representations of the neurons which are much faster to work with and can be used to compute various statistics such as the total length of the neuron.
For the public snapshots (630 and 783) we provide precomputed, high quality
skeletons for all proofread(!) neurons via get_skeletons():
>>> sk = flywire.get_skeletons(720575940625431866)
>>> sk
| type | navis.TreeNeuron |
|---|---|
| name | skeleton |
| id | 720575940625431866 |
| n_nodes | 3885 |
| n_connectors | None |
| n_branches | 764 |
| n_leafs | 859 |
| cable_length | 2236532.0 |
| soma | [467, 470, 472, 475, 477, 479, 480, 631, 635, ... |
| units | 1 nanometer |
These skeletons are also available for bulk download via this Github repository: https://github.com/flyconnectome/flywire_annotations
What if there is no precomputed skeleton available for your neuron(s) of interest?
In that case you have two options to generate skeletons yourself: 1. Generate a high-res skeleton from scratch (slow) 2. Generate a low-res skeleton using the L2 cache (fast)
Let’s first demo the high-res skeleton:
%%time
>>> # Generate a high-res skeleton from scratch
>>> sk = flywire.skeletonize_neuron(720575940625431866, progress=False)
>>> sk
CPU times: user 20.2 s, sys: 2.12 s, total: 22.3 s
Wall time: 27.4 s
| type | navis.TreeNeuron |
|---|---|
| name | None |
| id | 720575940625431866 |
| n_nodes | 10326 |
| n_connectors | None |
| n_branches | 740 |
| n_leafs | 854 |
| cable_length | 2491188.964376 |
| soma | 12908 |
| units | 1 nanometer |
As you can see skeletonization is an expensive process and it can take several minutes on large neurons. Our examples neuron took around 30s on a laptop which is about average.
If you need to skeletonize many neurons you can use
fafbsegb.flywire.skeletonize_neurons_parallel() to speed things up.
Also note that the default settings for the skeletonization work well enough for 9 out of 10 neurons but you might have to play around with them if the skeleton looks odd or takes very long to generate.
>>> fig, ax = navis.plot2d(sk, color="red")
>>> ax.azim = ax.elev = -90
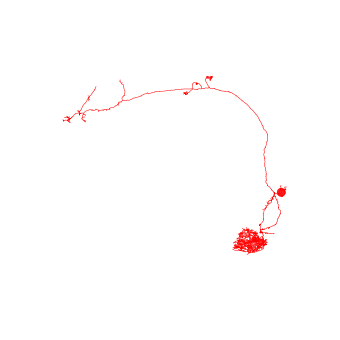
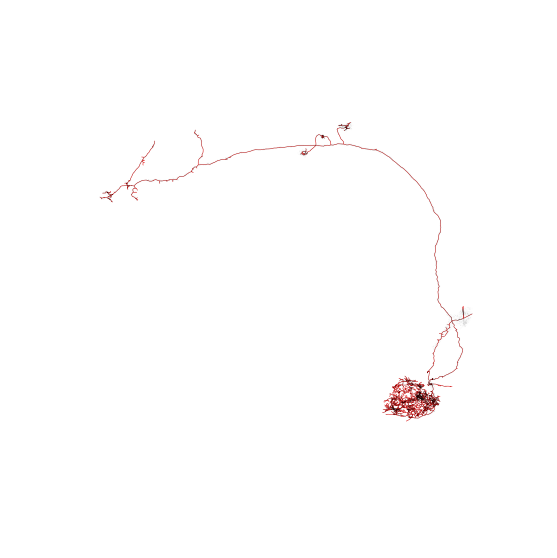
For comparison, lets plot mesh (transparent grey) and skeleton (red) side-by-side:
>>> fig, ax = navis.plot2d([sk, n], color=["r", (.3, .3, .3, 0.1)], figsize=(10, 10), method='3d_complex')
>>> ax.azim = ax.elev = -90
We can also use plot3d instead of plot2d. Try zooming in to see
how the skeleton compares to the mesh.
>>> navis.plot3d([sk, n], color=["r", (0.3, 0.3, 0.3, 0.3)])
If you don’t need high-res meshes or skeletons, you might want to take a look at on-the-fly L2 skeletons. These are coarser but much faster to generate:
>>> # Time how long it takes to grab an L2 skeleton
>>> %time l2_sk = flywire.get_l2_skeleton(720575940625431866)
CPU times: user 135 ms, sys: 9.33 ms, total: 144 ms
Wall time: 3.1 s
Note
For NBLASTs, there is also a function for on-the-fly dotprops:
>>> l2_dp = flywire.get_l2_dotprops(720575940625431866)
Let’s compare the full-res and L2 skeleton:
>>> fig, ax = navis.plot2d([sk, l2_sk], color=["r", "c"], method='3d_complex')
>>> ax.azim = ax.elev = -90
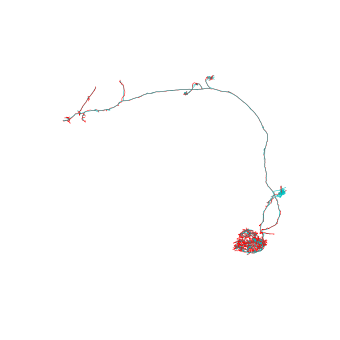
These L2 skeletons (or dotprops) are good enough for most morphological analyses and NBLAST!
Note
Have a look at the tutorials
for navis to learn more about what you can do with the neurons.