Fetching connectivity#
Some background#
Thanks to Buhmann et al., 2019 we have a synapse prediction for the FAFB dataset.
To facilitate querying that data for FlyWire, Sven Dorkenwald, Forrest Collman et al. have loaded
these data into their CAVE annotation backend. fafbseg.flywire provides a couple
convenient wrappers to fetch synapses and connectivity.
Here, we will focus on querying connectivity for the FlyWire project and for that a bit of background information is in order:
The synapse predictions are actually ~130M connections (220M before filtering) each of which consists of a presynaptic and a postsynaptic x/y/z coordinate plus some meta data. These coordinates have been mapped to FlyWire supervoxel IDs. The tricky (read: expensive/slow) part is mapping these static supervoxels to the ever changing root IDs. This step is being done by the “materialization” CAVE engine.
These materializations are run once every night and kept for a couple days before being discarded. Some versions - like the FlyWire public release a.k.a. materialization version 630 - are immortal.
The important thing to keep in mind is: each materialization represent a snapshot for the root IDs that existed at that time.
Let’s start with an example of how you can check the dates:
>>> from fafbseg import flywire
To illustrate let’s fetch the currently available materializations. Note that this will only work if you have production data access!
>>> flywire.get_materialization_versions(dataset="production")
| is_merged | datastack | id | status | version | time_stamp | expires_on | valid | |
|---|---|---|---|---|---|---|---|---|
| 0 | False | flywire_fafb_production | 823 | AVAILABLE | 833 | 2024-01-09 05:10:00 | 2024-01-11 04:10:00 | True |
| 1 | False | flywire_fafb_production | 822 | AVAILABLE | 832 | 2024-01-06 05:10:00 | 2024-01-13 04:10:00 | True |
| 2 | False | flywire_fafb_production | 820 | AVAILABLE | 830 | 2024-01-03 05:10:00 | 2024-02-07 04:10:00 | True |
| 3 | False | flywire_fafb_production | 813 | AVAILABLE | 823 | 2023-12-20 05:10:00 | 2024-01-17 04:10:00 | True |
| 4 | False | flywire_fafb_production | 773 | AVAILABLE | 783 | 2023-09-30 05:10:00 | 2121-11-10 07:10:00 | True |
| 5 | True | flywire_fafb_production | 619 | AVAILABLE | 630 | 2023-03-21 08:10:00 | 2121-11-10 07:10:00 | True |
| 6 | True | flywire_fafb_production | 560 | AVAILABLE | 571 | 2023-01-10 08:11:00 | 2121-11-10 07:10:00 | True |
| 7 | True | flywire_fafb_production | 515 | AVAILABLE | 526 | 2022-11-17 08:10:00 | 2121-11-10 07:10:00 | True |
| 8 | True | flywire_fafb_production | 247 | AVAILABLE | 258 | 2022-01-17 08:10:00 | 2121-02-18 08:10:00 | True |
Do the same for the public release - note that at the moment it contains only version 630.
>>> flywire.get_materialization_versions(dataset="public")
| is_merged | datastack | id | status | version | time_stamp | expires_on | valid | |
|---|---|---|---|---|---|---|---|---|
| 0 | True | flywire_fafb_public | 718 | AVAILABLE | 630 | 2023-03-21 08:10:00 | 2121-11-10 07:10:00 | True |
For the production dataset only: we can also project the latest materialization into the now and get a “live” version for the current root IDs.
As of version 1.14.0, fafbseg will try to automatically find a
materialization that contains your query root IDs. In general, however, it’s
good practive to make sure that your query root IDs match one of the existing
materialization versions.
Let’s use the public release data to explore this:
>>> flywire.set_default_dataset("public")
Default dataset set to "public"
Synapses#
OK, let’s start by fetching synapses for a given root ID
>>> syn = flywire.get_synapses(
... 720575940625431866,
... materialization="auto" # "auto" is the default value
... )
>>> syn.head()
Using materialization version 630
| pre | post | cleft_score | pre_x | pre_y | pre_z | post_x | post_y | post_z | id | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 720575940618561403 | 720575940625431866 | 144 | 479728 | 242036 | 23760 | 479856 | 242028 | 23760 | 56078503 |
| 1 | 720575940631121739 | 720575940625431866 | 63 | 475736 | 237908 | 21800 | 475716 | 237808 | 21760 | 111010940 |
| 2 | 720575940612003441 | 720575940625431866 | 142 | 437296 | 131392 | 185360 | 437408 | 131420 | 185400 | 183931595 |
| 3 | 720575940618654016 | 720575940625431866 | 144 | 348664 | 147136 | 162720 | 348688 | 147224 | 162720 | 24048637 |
| 4 | 720575940626518302 | 720575940625431866 | 129 | 485796 | 248296 | 28200 | 485840 | 248120 | 28280 | 3443470 |
Note how it says “Using materialization version 630” - that’s because this is the only available materialization for the public release. If you try to query root IDs that did not exist at this materialization it will complain!
We can also force a query against the live data. Note that this will be considerably slower than queries against a materialized table! Importantly, this only works with if you have access to the production dataset.
>>> syn_live = flywire.get_synapses(
... 720575940625431866, materialization="live", filtered=False, dataset="production"
... )
>>> syn_live.head()
| pre | post | cleft_score | pre_x | pre_y | pre_z | post_x | post_y | post_z | id | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 720575940614727903 | 720575940625431866 | 0 | 503800 | 205396 | 32840 | 503672 | 205416 | 32800 | 71828773 |
| 1 | 720575940633938349 | 720575940625431866 | 41 | 486888 | 246616 | 33240 | 486868 | 246484 | 33240 | 71914527 |
| 2 | 720575940631975884 | 720575940625431866 | 83 | 480208 | 244212 | 30080 | 480144 | 244384 | 30040 | 191440776 |
| 3 | 720575940617537570 | 720575940625431866 | 166 | 480960 | 242752 | 23840 | 481092 | 242724 | 23800 | 56078511 |
| 4 | 720575940613489617 | 720575940625431866 | 142 | 475384 | 235384 | 25960 | 475496 | 235376 | 25960 | 52231029 |
Using a root ID that has no materialization, or multiple root IDs that don’t have any materialization in common results in an error:
>>> # These are two root IDs for the same neuron (one old, one current)
>>> # THIS IS EXPECTED TO FAIL!
>>> syn = flywire.get_synapses([720575940618984129, 720575940620240833], materialization="auto")
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[10], line 3
1 # These are two root IDs for the same neuron (one old, one current)
2 # THIS IS EXPECTED TO FAIL!
----> 3 syn = flywire.get_synapses([720575940618984129, 720575940620240833], materialization="auto")
File ~/Google Drive/Cloudbox/Github/fafbseg-py/fafbseg/flywire/annotations.py:122, in parse_neuroncriteria.<locals>.outer.<locals>.inner(*args, **kwargs)
120 kwargs[key] = nc.get_roots()
121 try:
--> 122 return func(*args, **kwargs)
123 except NoMatchesError as e:
124 if allow_empty:
File ~/Google Drive/Cloudbox/Github/fafbseg-py/fafbseg/flywire/utils.py:166, in inject_dataset.<locals>.outer.<locals>.inner(*args, **kwargs)
164 if disallowed and ds in disallowed:
165 raise ValueError(f'Dataset "{ds}" not allowed for function {func}.')
--> 166 return func(*args, **kwargs)
File ~/Google Drive/Cloudbox/Github/fafbseg-py/fafbseg/flywire/synapses.py:466, in get_synapses(x, pre, post, attach, filtered, min_score, transmitters, neuropils, clean, materialization, batch_size, dataset, progress)
463 materialization = client.materialize.most_recent_version()
465 if materialization == "auto":
--> 466 materialization = find_mat_version(ids, dataset=dataset, verbose=progress)
467 else:
468 _check_ids(ids, materialization=materialization, dataset=dataset)
File ~/Google Drive/Cloudbox/Github/fafbseg-py/fafbseg/flywire/utils.py:707, in find_mat_version(ids, verbose, allow_multiple, raise_missing, dataset)
703 raise ValueError('Given root IDs do not co-exist in any of the available '
704 'materialization versions (including live). Try updating '
705 'root IDs and rerun your query.')
706 else:
--> 707 raise ValueError('Given root IDs do not co-exist in any of the available '
708 'public materialization versions. Please make sure that '
709 'the root IDs do exist and rerun your query.')
ValueError: Given root IDs do not co-exist in any of the available public materialization versions. Please make sure that the root IDs do exist and rerun your query.
See the last line in the error message? It tells you that these IDs never co-existed and hence can not be queried together.
Instead of using “auto” you can also provide a specific materialization version. In that case you will get a warning if your queries don’t match the materialization but it won’t throw an exception:
>>> syn = flywire.get_synapses(720575940620240833, materialization=630)
>>> syn.head()
Some root IDs were already outdated at materialization 630 and synapse/connectivity data will be inaccurrate: 720575940620240833 Try updating the root IDs using flywire.update_ids or flywire.supervoxels_to_roots if you have supervoxel IDs, or pick a different materialization version.
| pre | post | cleft_score | pre_x | pre_y | pre_z | post_x | post_y | post_z | id |
|---|
Note that in this example we got an empty dataframe because the root ID didn’t exist at that specific materialization!
See fafbseg.flywire.is_latest_root() and fafbseg.flywire.update_ids()
for mapping root IDs to materializations:
>>> # Search for this neuron's ID at materialization 630 (public release version)
>>> new_id = flywire.update_ids(720575940620240833, timestamp="mat_630")
>>> new_id
| old_id | new_id | confidence | changed | |
|---|---|---|---|---|
| 0 | 720575940620240833 | 720575940622894616 | 0.991647 | True |
With the new up-to-date root ID, the query will work as expected:
>>> syn = flywire.get_synapses(720575940622894616)
>>> syn.head()
Using materialization version 630
| pre | post | cleft_score | pre_x | pre_y | pre_z | post_x | post_y | post_z | id | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 720575940627172100 | 720575940622894616 | 142 | 384712 | 159552 | 184160 | 384840 | 159424 | 184200 | 166252986 |
| 1 | 720575940634147936 | 720575940622894616 | 135 | 386928 | 156476 | 181800 | 386904 | 156556 | 181840 | 155850631 |
| 2 | 720575940632826925 | 720575940622894616 | 144 | 341452 | 147360 | 173040 | 341424 | 147504 | 173040 | 228310877 |
| 3 | 720575940608970197 | 720575940622894616 | 116 | 351400 | 163320 | 187320 | 351492 | 163280 | 187360 | 66153949 |
| 4 | 720575940624632247 | 720575940622894616 | 145 | 358000 | 145404 | 163080 | 357864 | 145396 | 163080 | 87457018 |
Alternatively: if you keep track of your neurons via x/y/z coordinates or
supervoxel IDs, you can also use the timestamp parameter in e.g.
fafbseg.flywire.supervoxels_to_roots() to get the root IDs at the same
time as one of the materializiations. This can be slightly faster than live
queries.
Neurons & synapses#
You find yourself wanting to associate synapses with the neuron’s skeleton or mesh - e.g. for plotting or analyses. Here’s how you would do that:
>>> # Fetch a skeleton
>>> sk = flywire.get_skeletons(720575940625431866)
>>> # The skeleton does not yet have any synapse data associated with it
>>> sk.connectors
To fetch the neuron’s synapses and attach them to the skeleton you can do this:
>>> flywire.get_synapses(sk, attach=True)
>>> # Now the neuron has a connector table
>>> sk.connectors.head()
Using materialization version 630
| connector_id | x | y | z | cleft_score | partner_id | type | node_id | |
|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 378580 | 141040 | 176720 | 151 | 720575940486790143 | pre | 324 |
| 1 | 1 | 448544 | 121840 | 192120 | 161 | 720575940617293396 | pre | 706 |
| 2 | 2 | 449832 | 122628 | 195640 | 157 | 720575940659323009 | pre | 704 |
| 3 | 3 | 446608 | 124216 | 195760 | 151 | 720575940659323009 | pre | 652 |
| 4 | 4 | 361652 | 129832 | 159520 | 61 | 720575940532780653 | pre | 265 |
Let’s illustrate by plotting the skeleton plus its synapses:
>>> import navis
>>> # Presynapses are red; postsynapses are light blue
>>> fig, ax = navis.plot2d(sk, connectors=True, color='lightgrey')
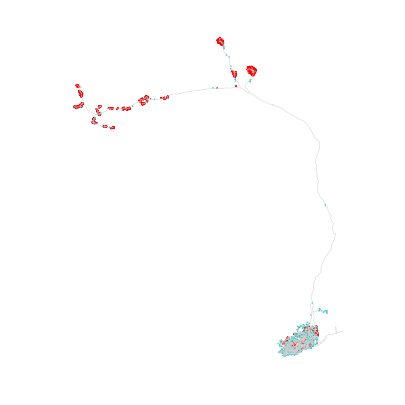
Connections#
Above you learned how to fetch individual synapses. You could distil those into an edge list by aggregating over the pre- and postsynaptic root IDs but that’s rather tedious. Fortunately, there is an easier (and faster) way of doing it:
>>> # These are current root IDs of RHS DA1 uPNs
>>> da1_roots = [
... 720575940604407468,
... 720575940623543881,
... 720575940637469254,
... 720575940617229632,
... 720575940621239679,
... 720575940623303108,
... 720575940630066007,
... ]
>>> # Get the partners for these neurons
>>> edge_list = flywire.get_connectivity(da1_roots)
>>> edge_list.head()
Using materialization version 630
| pre | post | weight | |
|---|---|---|---|
| 0 | 720575940611174702 | 720575940630066007 | 104 |
| 1 | 720575940611174702 | 720575940621239679 | 89 |
| 2 | 720575940619061662 | 720575940630066007 | 81 |
| 3 | 720575940611174702 | 720575940637469254 | 78 |
| 4 | 720575940611174702 | 720575940604407468 | 75 |
Note
You can also use NeuronCriteria to work with
annotations instead of passing root IDs directly. See the
annotation tutorial for details.
Generate a neuroglancer link with the top 30 downstream partners:
>>> # Grab the top 30 downstream partnes
>>> top30ds = edge_list[edge_list.pre.isin(da1_roots)].iloc[:30]
>>> # Generate a URL with these partners
>>> flywire.encode_url(segments=top30ds.post.values, open=False)
'https://ngl.flywire.ai/?json_url=https://globalv1.flywire-daf.com/nglstate/5912498324635648'
We can also generate an adjacency matrix:
>>> adj = flywire.get_adjacency(sources=da1_roots, targets=da1_roots)
>>> adj
Using materialization version 630
| target | 720575940604407468 | 720575940623543881 | 720575940637469254 | 720575940617229632 | 720575940621239679 | 720575940623303108 | 720575940630066007 |
|---|---|---|---|---|---|---|---|
| source | |||||||
| 720575940604407468 | 0.0 | 2.0 | 1.0 | 2.0 | 1.0 | 1.0 | 1.0 |
| 720575940623543881 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 2.0 | 0.0 |
| 720575940637469254 | 0.0 | 2.0 | 0.0 | 3.0 | 0.0 | 1.0 | 1.0 |
| 720575940617229632 | 1.0 | 2.0 | 1.0 | 0.0 | 2.0 | 1.0 | 1.0 |
| 720575940621239679 | 6.0 | 1.0 | 1.0 | 5.0 | 0.0 | 0.0 | 2.0 |
| 720575940623303108 | 0.0 | 1.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 |
| 720575940630066007 | 0.0 | 4.0 | 0.0 | 1.0 | 3.0 | 0.0 | 0.0 |
>>> import seaborn as sns
>>> ax = sns.heatmap(adj, cmap="Blues", square=True)

I hope this tutorial gave you a flavor and some entry points to start using these data. Please see the API reference for a full list of connectivity-related functions.