Transforming data between FAFB14 and FlyWire#
Units matter!#
Before we get started there is one very important thing to keep in mind:
Spatial data can be in physical space (e.g. microns or nanometers) or in voxels. For FAFB, you will typically find stuff to be in either nanometers or in 4x4x40nm voxels. Some simple rules of thumb:
Data (neurons, connectors, etc.) in CATMAID are in nanometers
Data (neuron meshes) in FlyWire are in nanometers
The CATMAID interface shows both physical units [nm] and voxels
The FlyWire neuroglancer interface typically uses voxels
Bottom line: you need to be aware of the units of the object you are trying to transform.
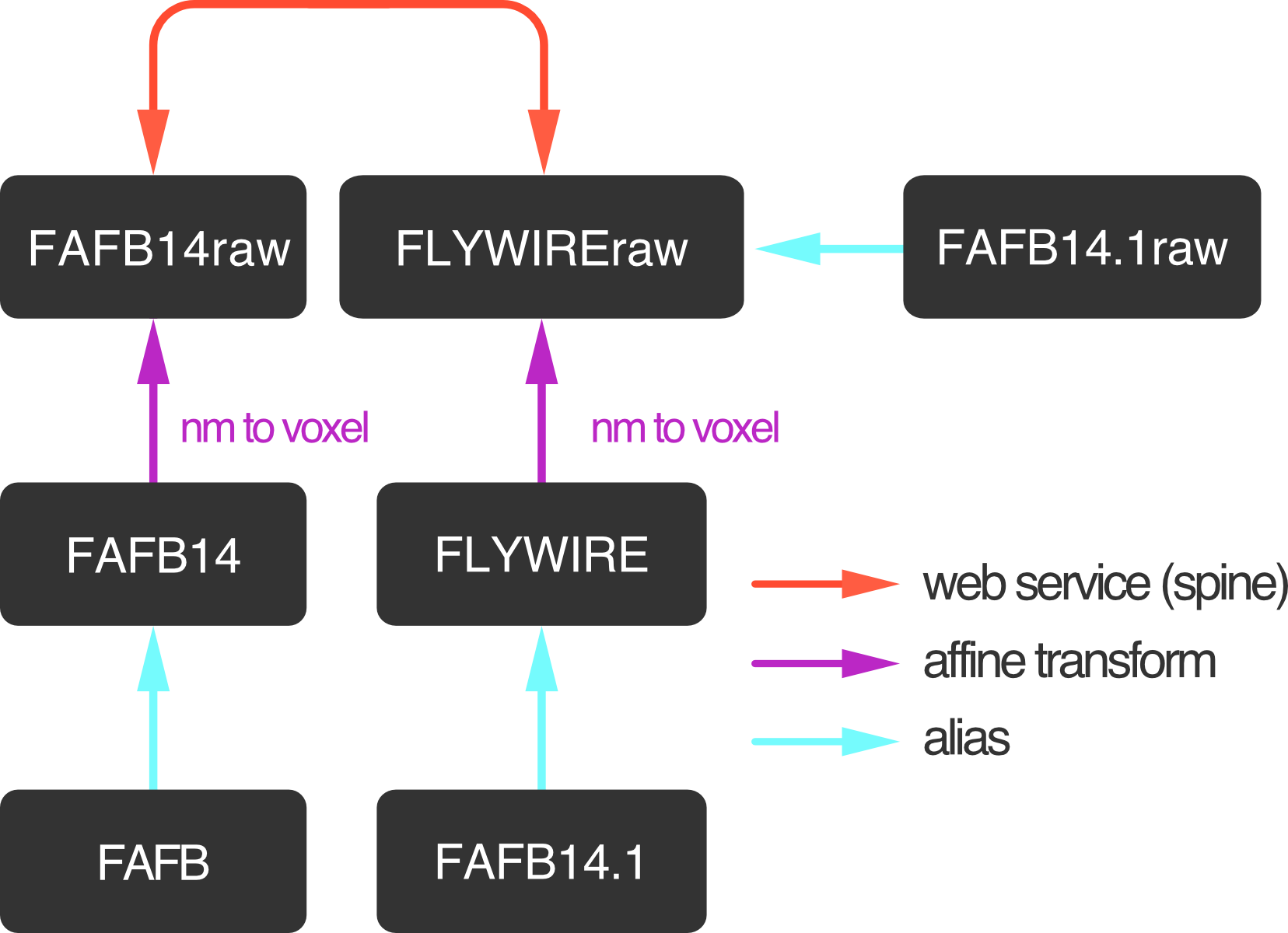
Transforming coordinates#
There are two ways to transform spatial data (neurons, meshes, x/y/z coordinates)
between FAFB14 and FlyWire space (also called “FAFB14.1”). The most convenient way
is to use navis built-in transform system.
As soon as you import fafbseg, the following transforms become available to navis:
Let’s take it for a spin:
>>> import fafbseg
>>> import navis
First example: map a single location from FAFB14 to FlyWire.
In CATMAID, left-click on the little pin icon in the upper right (next to the `Pos, Id, ` field) to copy the current location (in nanometers) to the clipboard. Note that holding shift while clicking on the icon copies the location in voxels.
>>> import numpy as np
>>> # The location from the clipboard
>>> pos = np.array([[249960, 184691.375, 174240]])
Looking at above bridging graph, our coordinates are in "FAFB14" (nm) space
and we want to go to "FLYWIREraw" (voxel) so that we can copy-paste the
coordinates into the FlyWire neuroglancer”:
>>> navis.xform_brain(pos, source="FAFB14", target="FLYWIREraw")
Transform path: FAFB14 -> FAFB14raw -> FLYWIREraw
array([[62648.5 , 46291.34375, 4356. ]])
If you copy the above coordinates into the FlyWire neuroglancer, they should get you exactly to the same location.
The advantage of navis.xform_brain is that it ties in with other available
transforms. So in theory, you could go from FLYWIRE all the way to
JRC2018F with a command.
The alternative to navis.xform_brain is to use a lower-level function:
fafbseg.xform.fafb14_to_flywire(). This function and its inverse
counterpart fafbseg.xform.flywire_to_fafb14() give you a bit more
control over the transform.
>>> fafbseg.xform.fafb14_to_flywire(pos, coordinates="nm")
array([[250594. , 185165.38, 174240. ]], dtype=float32)
Note that these lower-level functions always return in the same space - hence above result is also in nanometers.
Second example: transforming a neuron
>>> import pymaid
>>> # Connect to the VirtualFlyBrain's public CATMAID instance
>>> cn = pymaid.CatmaidInstance(
... server="https://fafb.catmaid.virtualflybrain.org/", project_id=1, api_token=None
... )
>>> # Load a neuron
>>> n = pymaid.get_neuron(16)
>>> # Xform the neuron - note we stay in nanometer space for now
>>> xf = navis.xform_brain(n, source="FAFB14", target="FLYWIRE")
INFO : Global CATMAID instance set. Caching is ON. (pymaid)
Transform path: FAFB14 -> FAFB14raw -> FLYWIREraw -> FLYWIRE
>>> # Co-visualize the original (FAFB space) and the transformed (FlyWire space) neurons
>>> fig, ax = navis.plot2d([n, xf], color=["r", "c"], lw=0.5, method='3d_complex')
>>> ax.azim = ax.elev = -90
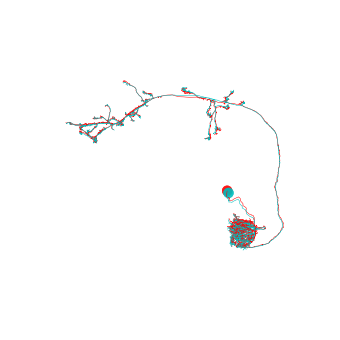
As you can see, the offsets are rather small - probably most obvious in the soma tract.